Is there a bias after DNA fragmentation?
Cutting DNA into small fragments is a key preparation step for DNA sequencing with NGS technology. To reduce errors and increase reliability of the sequence information, every genomic region should be sequenced several times. This means that several copies of a target DNA have to be cutted in different ways to produce overlapping fragments to ensure an good coverage of the whole region of interest. This approach is based on the general idea that genomic DNA break-points are random and sequence-independent.
The fragmentation bias
The Problem: DNA fragmentation is often non-random, especially for mechanical methods like ultrasonication shearing (1). Several studies showed that this non-random cleavage is the result of sequence-dependent conformational dynamics (1,2,3). These dynamics are likely modulated by the intensity of the sugar ring S↔N interconversion (2,4). GC-content and epigenetic mechanisms based on d(CpG) methylation might also influence the fragmentation process (5).
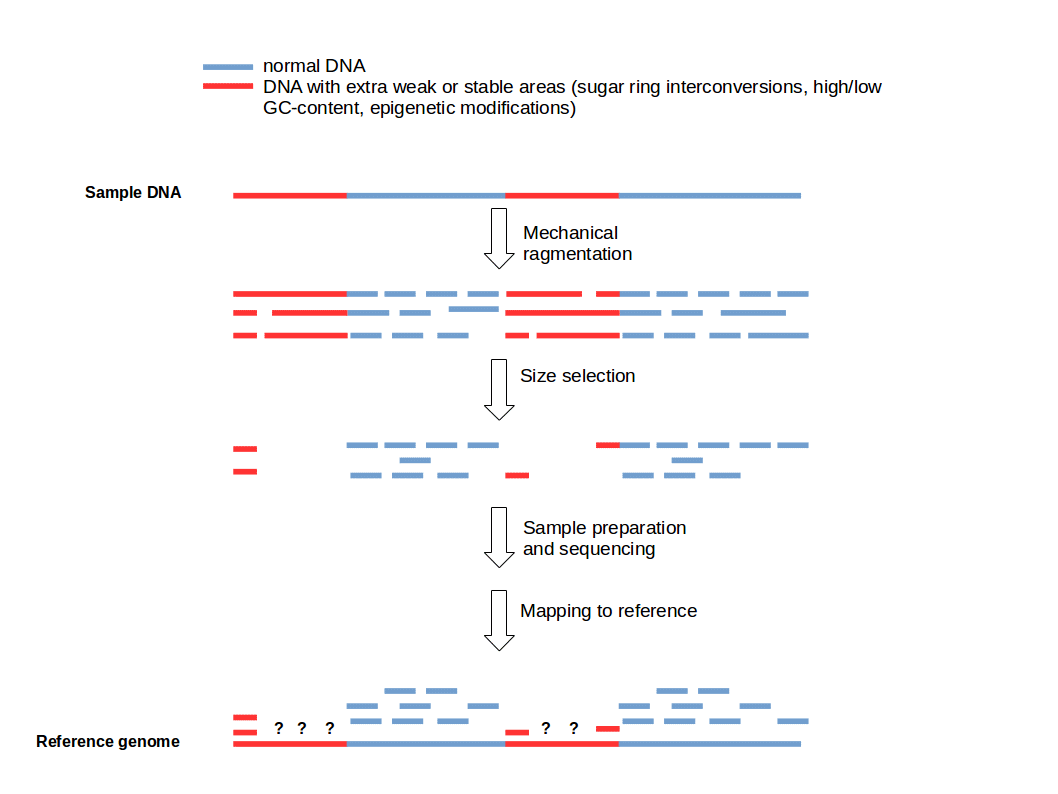 Figure 1: Fragmentation Bias. Weak and strong DNA areas creating predetermined breaking points resulting in low coverage for some sequenced DNA regions.
Figure 1: Fragmentation Bias. Weak and strong DNA areas creating predetermined breaking points resulting in low coverage for some sequenced DNA regions.
Consequences and improvement
The fragmentation bias leads to non-uniform coverage of various genomic regions (Fig. 1). It depends on various properties of the used DNA (e.g. GC-content) and the applied fragmentation method (physical, enzymatic, and chemical) whereby fragmentation methods based on the action of the hydrodynamic forces on DNA, produce similar biases (1). For some fragmentation methods it is possible to lower the systematic bias. For example ultrasonication shearing can be improved by addition of particular metallic ions (e.g. Ag+) (6).
This shows you that DNA unfortunately cannot be fragmented full uniformity. Compare this to cooking spaghetti. Imagine some parts of the spaghetti are already parboiled and flexible and will not break as easy as the uncooked and hard regions. The fragmentation bias affects especially mechanical methods like ultrasonication. Consider that there are alternative fragmentation methods based on chemical or biological reactions that you can use alternatively or additionally.
References:
- Poptsova, M. S. et al. Non-random DNA fragmentation in next-generation sequencing. Sci. Rep. 4, (2014).
- Grokhovsky, S. L. et al. Sequence-Specific Ultrasonic Cleavage of DNA. Biophys. J. 100, 117–125 (2011).
- Grokhovsky, S. L. Specificity of DNA cleavage by ultrasound. Mol. Biol. 40, 276–283 (2006).
- Il’icheva, I. A., Nechipurenko, D. Y. & Grokhovsky, S. L. Ultrasonic cleavage of nicked DNA. J. Biomol. Struct. Dyn. 27, 391–398 (2009).
- Lazarovici, A. et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U. S. A. 110, 6376–6381 (2013).
- Grokhovsky, S. L. in Ultrasonics: Theory, Techniques and Practical Applications (eds. Ayabito, H. & Katsukawa, M.) (2013).
Would you like to sharpen your NGS data analysis skills?
Join one of our public workshops!About us
ecSeq is a bioinformatics solution provider with solid expertise in the analysis of high-throughput sequencing data. We can help you to get the most out of your sequencing experiments by developing data analysis strategies and expert consulting. We organize public workshops and conduct on-site trainings on NGS data analysis.
Last updated on April 20, 2017