How to analyze NGS data: An overview of nine different IT solutions
Have you been given the task to work with Next-Generation Sequencing (NGS) data? Before you start and bind yourself to any existing software or online platform, you might want to be familiar with the options available on the market. Nowadays, there is such a broad range of different solutions available, that it is worth comparing them before starting any project. This post aims to give a first taxonomy of the crowded space of IT solutions for NGS data analysis.
Note: This article focuses on software solutions. The usage of these tools requires some understanding of the involved bioinformatics methods. You have to be able to interpret the results properly and spot data analysis issues yourself. The alternative is to rely on NGS analysis services offered by bioinformatics providers or sequencing providers, which will not be discussed here.
To cloud, or not to cloud
The first important decision usually is whether you are willing to use, or maybe prefer to use, a cloud-based solution for your data analysis. The obvious benefit of having both computation and data in the cloud is that you do not have to take care of local computing and storage resources yourself - which of course only works when all the data and needed workflows are available in the cloud. Also pay attention to existing organizational policies that might put any cloud-based solution out of the question for you.
Overview
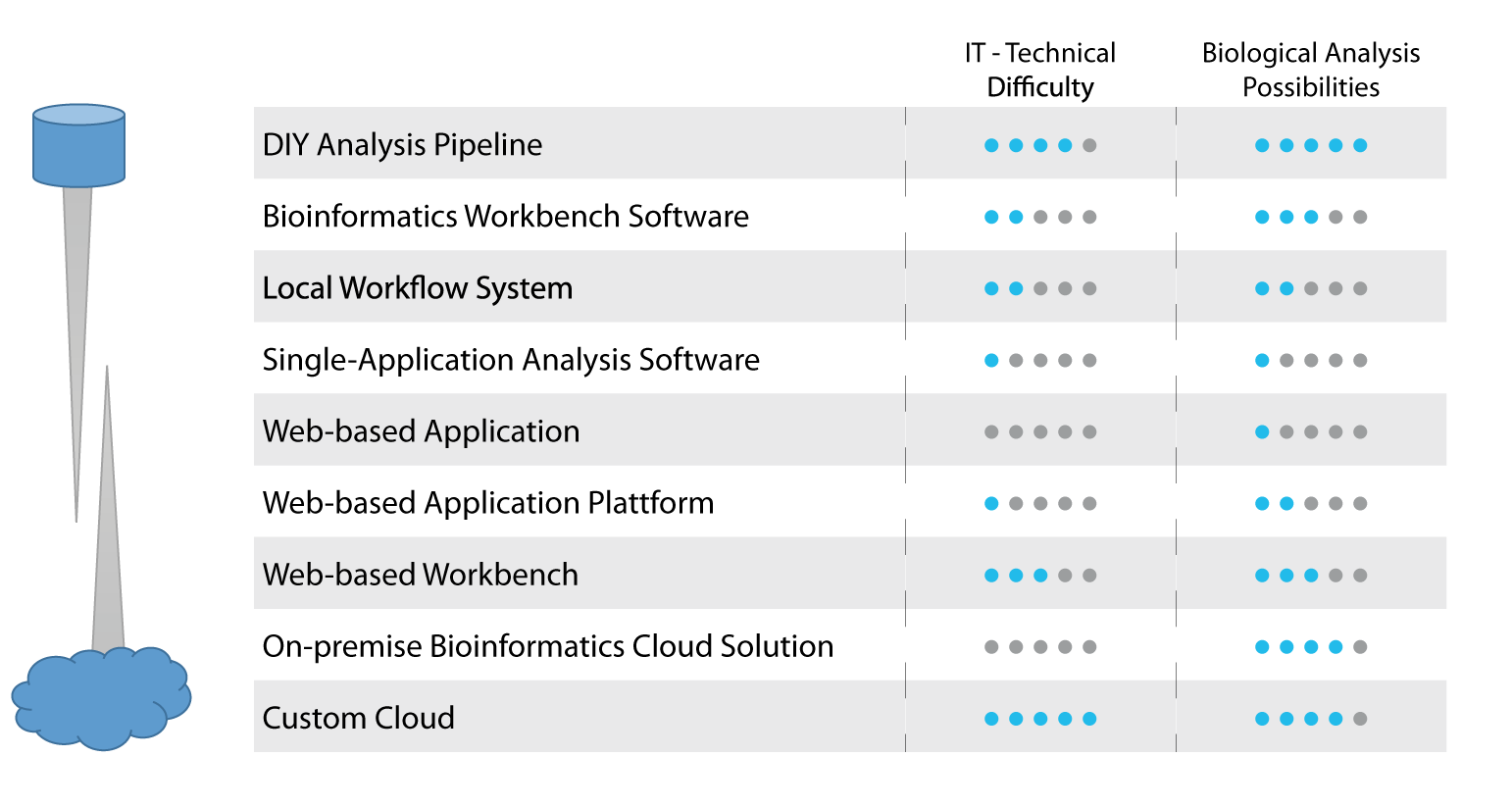
The following infographic gives an overview over the different solutions which will be described in more detail below. We have also indicated in that picture how these solutions, in our opinion, differ in two important aspects. Firstly, IT/technical difficulty describes the level of expertise in IT and NGS bioinformatics needed to setup these systems and in using them to get to reliable results. Secondly, biological analysis possibilities refers to the extent and flexibility of the solution to answer also particular (off-the-shelf) biological questions. The second point is important, as an analysis oftentimes is not finished after one single step, e.g. the result of a DNA variant calling is itself not sufficient but needs to be enriched with biomedical information.
DIY Pipeline
Today, this can safely be considered as the default solution for analyzing NGS data: combine available open-source bioinformatics tools with your own scripts, in order to implement a custom workflow for your current data analysis problem. Luckily there is quite a number of NGS-related bioinformatics tools (read aligners, variant callers, adapter trimmers, etc.) out there. With a good understanding of the algorithms, specifications and characteristics of every single tool, one can develop a solution for almost all tasks. Tailor these to your infrastructure and batch processing systems as needed.
Bioinformatics Workbench Software
These standalone desktop applications offer a broad range of biological data analysis and visualization features. Their main advantage is user-friendliness. These all-in-one bioinformatics suites allow you to do both secondary analysis and various downstream analysis tasks using the same graphical user interface. But, as for all local software solutions, their ability to deal with NGS data is limited to the processing power of the computer the software is running on. Compared to the freedom of DIY pipelines, you are limited to the tasks the workbench solution offer.
Local Workflow System
These software systems can be installed within your internal network. They offer an easy way to run a specific set of analysis protocols coupled with extra features, such as high scalability data processing, experiment management, integration of external data sources and result annotation. These applications are typically accessed using a web-based interface rather than using desktop applications.
Single-Application Analysis Software
A standalone software developed for one specific task, such as microbial genome assembly or plant gene expression analysis. This focus allows the developers of the software to design it for specific hardware requirements and implement a range of features that are relevant for exactly this application.
Web-based Application
This refers to solutions that provide a web-based service for specific NSG analyses. The most important goal is to make it as easy as possible to carry out a certain analysis (“push-button analysis”) and provide extended features that make sense only for a specific taxon/analysis/protocol. The most famous of these are the online variant analysis services (“GATK online”).
Web-based Application Platform
The logical extension of the singleton online service is the web-based platform providing various NGS analyses via “Apps”. Again, each “App” runs a very specific computational protocol on the data. Ideally, the output of one app can be the input of another app, thus allowing you to do also certain downstream analyses within the platform. Additional features include storage, data and experiment management and result sharing.
Web-based Workbench
This is the web-based analog to the standalone workbench software. It gives you access to a larger number of individual tools and analysis tasks which can be then combined to larger workflows. Collaboration features allow to share data, results and workflows with partners that have access to the system.
On-premise bioinformatics cloud solution
This is a variant of the cloud-based bioinformatics platform where the provider allows arbitrary data analysis workflows to be included in their system. These are complemented by data management and collaboration features. They provide multiple ways to transfer data and interact with the computing environment.
Custom Cloud
Custom cloud means setting up a own analysis solution on one of the many cloud service providers. This usually involves setting up a computing cluster and a connected storage. There are images available that allow you to run some of the better known NGS tools without having to do tedious installation routines. Once everything is set up, you can run all of the analyses that you would run on a local cluster. Note that all intermediate data needs to be transferred through the internet to your local computer.
Discussion
Although the number of options seems large, we observe that many teams have to rely on custom solutions. This is due to the fact that the applications of sequencing are so diverse, that it is most of the time impossible to cover all needed analysis steps and fulfill all requirements.
Disclaimer: In our NGS analysis trainings, we try to use only free open source software (FOSS).
Would you like to sharpen your NGS data analysis skills?
Join one of our public workshops!About us
ecSeq is a bioinformatics solution provider with solid expertise in the analysis of high-throughput sequencing data. We can help you to get the most out of your sequencing experiments by developing data analysis strategies and expert consulting. We organize public workshops and conduct on-site trainings on NGS data analysis.
Last updated on October 07, 2016