Do you have two colors or four colors in Illumina?
The four-dye system, with four distinct ‘colors’ for calling the DNA bases, is the standard for Illumina technology. But lately Illumina changed it. For the new machines, like MiniSeq, NextSeq and NovaSeq, Illumina uses an alternative system with just two dyes. How is it possible to detect four different bases on that system?
The four-dye system
In the standard system, each of the four nucleotides is labeled with a separate dye. In each amplification cycle these dye signals are imaged by using two lasers and four filters (to allow discrimination between two-colors per laser). This system is also called 4-channel SBS (sequencing by synthesis), shown in Fig. 1 (left).
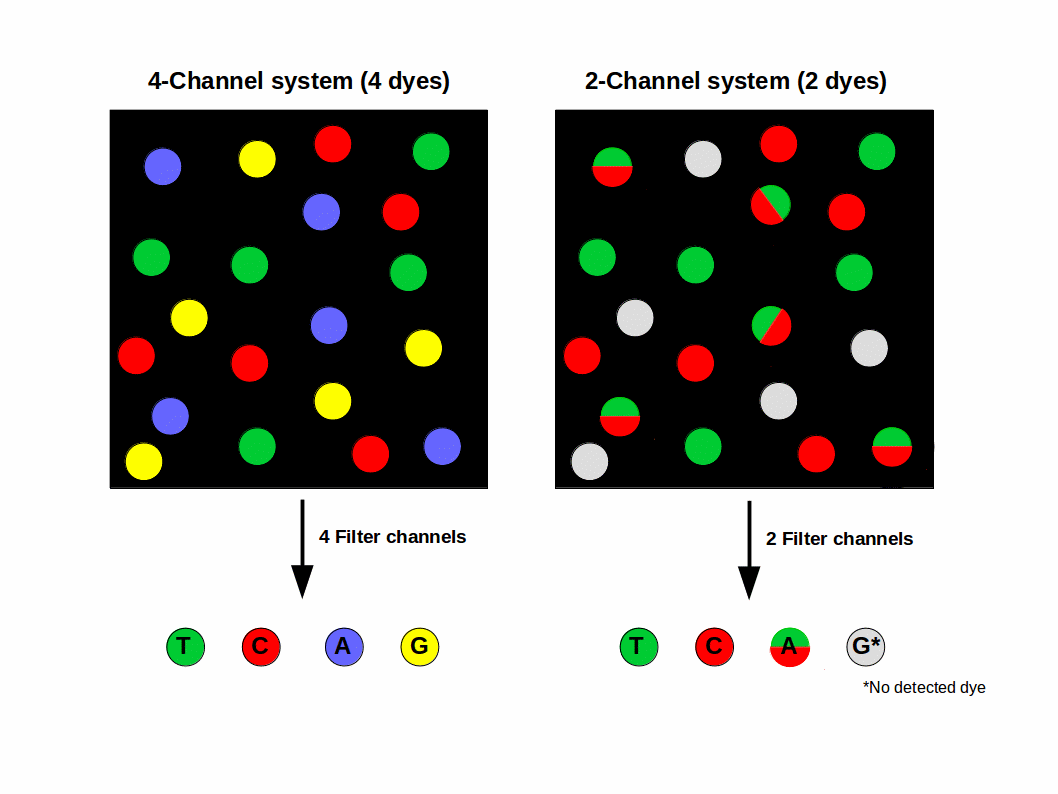 Figure 1: Comparison of 4- and 2-Channel system.
Figure 1: Comparison of 4- and 2-Channel system.
How to get the four bases with two colors?
Unlike the 4-channel system, 2-channel sequencing uses a mix of two dyes only, for example ‘red and green’. Clusters seen in red channel are interpreted as C whereas the ones seen in the green will be T bases. On the other hand, clusters observed in both red and green images will be flagged as A bases while unlabeled clusters (no dye) are identified as a G base (Fig. 1, right).
This system is called 2-channel SBS sequencing. Its strategy makes the real-time analysis quicker because the filter wheel has to change the emission filter just two times per run (2-Channel System) instead of four times (4-Channel System).
Downsides
The 2-channel System is a further development of the 4-channel system resulting in a faster sequencing process. Unfortunately it also has disadvantages: The base quality is determined, among other factors, by the purity of the emitted light signal of each cluster per run. Furthermore, incorrect base calls because of phasing lead to a rising pollution of the light signals over time, making it more difficult to differentiate the bases and to interpret the base quality. For example, the 2-channel system interprets a mix out of two light signals (red and green) as signal for adenine. Such a mixed red-green signal should always be adenine meaning that a red-green mixed signal due to phasing (and not by adenine) could be overlooked. This can distort base qualities and even the accuracy of the sequences.
Update: A Simplified Approach to Detecting DNA Bases Using a Single Fluorescent Dye
The 1-channel system in Illumina sequencing represents a further simplification of the detection procedure. Instead of using several dyes, all four DNA bases (A, T, C, G) are labelled with the same dye. The distinction between nucleotides is made by detecting the order of their incorporation into the growing DNA strand, rather than relying on different colours or colour combinations. This simplifies the sequencing process by reducing optical complexity, resulting in faster and more cost-effective data acquisition without compromising accuracy.
Would you like to sharpen your NGS data analysis skills?
Join one of our public workshops!About us
ecSeq is a bioinformatics solution provider with solid expertise in the analysis of high-throughput sequencing data. We can help you to get the most out of your sequencing experiments by developing data analysis strategies and expert consulting. We organize public workshops and conduct on-site trainings on NGS data analysis.
Last updated on February 27, 2017